Tumor Immunology
Prof. Dr. Dirk Jäger
We specialize in investigating tumor-immune interactions, creating immunotherapies and predictive biomarkers. The "PROMISE" trial studies stage IV lung cancer patients, analyzing samples with various techniques such as sequencing, spectroscopy, and cytometry. Our immunobioinformatic platform predicts neoepitopes, while collaboration projects "PIKT" and "TCR POC" focus on T cell receptors. Multiomics and clinical data integration is facilitated by a graph database.
Research Strategy
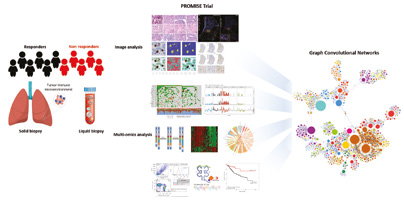
The exploratory biomarker trial “PROMISE - PRedictability of Outcome based on iMmunological sIgnatureS in lung cancEr – an explorative investigation”, a collaborative study at the Thoraxklinik, the DKFZ and the Heidelberg University Hospital, aims at identifying predictive biomarkers and signatures for response and resistance to standard of care treatment in stage IV lung cancer (NSCLC) patients. Tissue, serum, plasma, PBMCs, breath condensates and stool samples are collected longitudinally and will be analyzed by, e.g., WGS/WES, PanelSeq, RNASeq, Nanostring, Metagenomics, mass spectrometry, RAMAN spectroscopy and flow cytometry. Apart from NGS (Charoentong et al., 2017) and multiplex cytokine/chemokine analyses, tissue samples are explored by immunohistochemistry, virtual microscopy and image analysis. For the prediction of MHC I and II neoepitopes based on NGS data, a comprehensive immunobioinformatic platform has been set up. This platform is also applied in the multidisciplinary collaboration projects “PIKT” and “TCR POC” for the identification and validation of specific T cell receptors (TCR) targeting neoepitopes in solid tumors.
For an in-depth understanding of the “individual disease” and for the identification of predictive biomarkers, an integrated analysis of the multiomics and wet lab data as well as clinical data is performed. The integrated data analysis is supported by a recently established graph database.

Prof. Dr. Dirk Jäger
- +49 (0)6221 56-38590
- inka.zoernig@nct-heidelberg.de
Selected Publications
More information can be found here
Valous NA, Moraleda RR, Jäger D, Zörnig I, Halama N
Semin Immunol. 2020
Wells DK, van Buuren MM, Dang KK, Hubbard-Lucey VM, Sheehan KCF, Campbell KM, Lamb A, Ward JP, Sidney J, Blazquez AB, Rech AJ, Zaretsky JM, Comin-Anduix B, Ng AHC, Chour W, Yu TV, Rizvi H, Chen JM, Manning P, Steiner GM, Doan XC; Tumor Neoantigen Selection Alliance; Merghoub T, Guinney J, Kolom A, Selinsky C, Ribas A, Hellmann MD, Hacohen N, Sette A, Heath JR, Bhardwaj N, Ramsdell F, Schreiber RD, Schumacher TN, Kvistborg P, Defranoux NA
Cell. 2020
Kather JN, Pearson AT, Halama N, Jäger D, Krause J, Loosen SH, Marx A, Boor P, Tacke F, Neumann UP, Grabsch HI, Yoshikawa T, Brenner H, Chang-Claude J, Hoffmeister M, Trautwein C, Luedde T
Nat Med. 2019